a.)
a.) |
 b.)
b.) |
 c.)
c.) |
Phylogeny in the Red Ironbarks
(Eucalyptus series Siderophloiae)
By Pierre-Ulric Achour
Dec 2002 - Jan 2003
Supervisor: Maurice McDonald, CSIRO Forestry and Forest Products.
Introduction
Eucalyptus represents a large, taxonomically difficult genus. There are numerous groups in the genus that display complex variation patterns, which often make identification at species level difficult. One such group is the Red Ironbarks from the series Siderophloiae Blakely (Eucalyptus section Adnataria). This series includes over 20 species, which are characterised by deeply, longitudinally furrowed, kino-impregnated bark, terminal compound inflorescences with buds shedding their outer operculum early and fertile stamens irregularly flexed inside the bud (see Figure 1).
Most Red Ironbark species occur on the western plains and slopes of the Great Dividing Range in eastern Australia. Due to the high quality of their wood, large volumes of sawlogs are harvested from a number of species from natural stands, particularly in Queensland.
From a conservation and domestication perspective, the complex represents an important component of our native tree biodiversity.
Two widespread species of Red Ironbark – E. crebra (Narrow-Leaved Red Ironbark) and its close relative E. drepanophylla (Queensland Grey Ironbark) – have long been regarded as highly polymorphic, warranting critical taxonomic revision. Tony Bean, a taxonomist with the Queensland Herbarium, has a draft taxonomic revision of the Red Ironbark complex in which he proposes to split numerous new taxa from E. crebra and E. drepanophylla. His revision is the result of extensive fieldwork and examination of herbarium specimens undertaken over the past decade.
Experimental rationale and methodology
This study aimed to assess phylogenetic relationships among taxa in the Red Ironbark series Siderophloiae using allozyme markers, in order to reconcile current taxonomic species concepts with genetic evidence.
Plant material and sampling
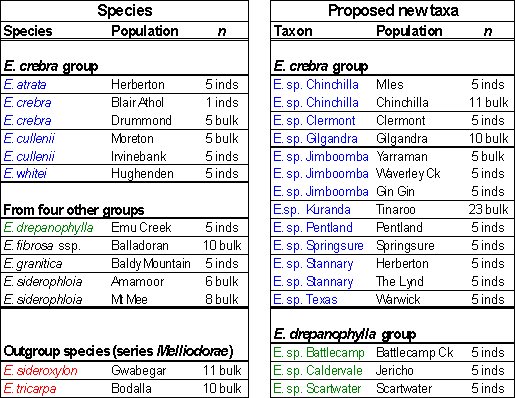
The study was based mainly on seed lots collected by Tony Bean and Maurice McDonald. It focused on the phylogeny of 13 taxa in the ‘E. crebra group’, which included nine proposed new taxa. Four taxa from the ‘E. drepanophylla group’ (with three proposed new taxa), the ‘E. siderophloia group’ (one taxon), the ‘E.granitica group’ (one taxon) and the ‘E. fibrosa group’ (one taxon) were also represented. Eucalyptus sideroxylon and E. tricarpa were included as outgroup species from the series Melliodorae. (See Table 1)
In total, twenty-nine populations were studied, representing eight described
species, 12 proposed new taxa and two outgroup species. These were collected
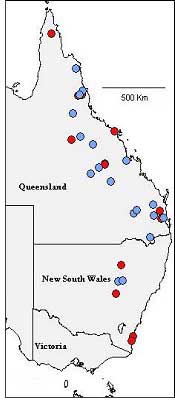
over an important distribution range, as displayed in Figure 2.
Table 1. Taxa and populations sampled for the study and number of trees representing each population.
Taxa arranged in groups according to an informal classification by Tony Bean: ‘E. crebra group’ in blue, ‘E. drepanophylla group’ in green and outgroups in red.; inds = seeds assayed from separate individual trees; bulk = seeds from mother trees mixed together.
Figure 2. Locations of populations studied. Known species are in red, proposed taxa are in blue.
Allozyme analysis
Twenty-five progeny from each population were grown for 8-10 days before being assayed. From a total of 16 enzyme systems tested, nine polymorphic loci were resolved from five enzyme systems (Aat, Gpi, Mdh, Pgd, Ugp). These loci were consistently scoreable while all others were either invariant or poorly resolved. A mean of seven alleles per locus was detected for the nine loci.
The BIOSYS program (Swofford & Selander 1981) was used to estimate allele frequencies for each population. Two programs for genetic distance matrix data from PHYLIP (Felsenstein 1995) were used to produce dendrograms. The program FITCH was used to create a tree using the Fitch-Margoliash and Least-squares distance method based on 500 bootstraps. In this method an evolutionary clock is not assumed and its estimates are based on unequal rates of change among different lineages (Weir 1996). A second tree was produced, also based on 500 bootstraps, using the program KITSCH. This program also uses the Fitch-Margoliash and Least-squares distance method but produces a tree typology comparable to the UPGMA method as it assumes a constant or equal rate of change is operative along all branches in the tree (Weir 1996).
Results and summary of findings
The dendrograms based on the Fitch-Margoliash and Least-squares
distance method are shown in Figure 3.
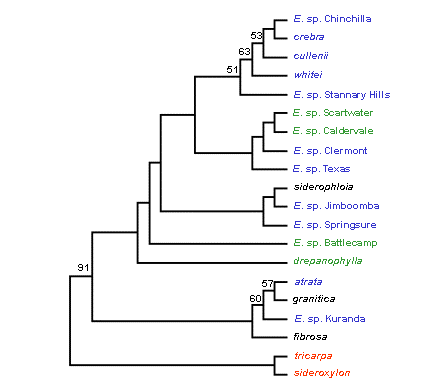
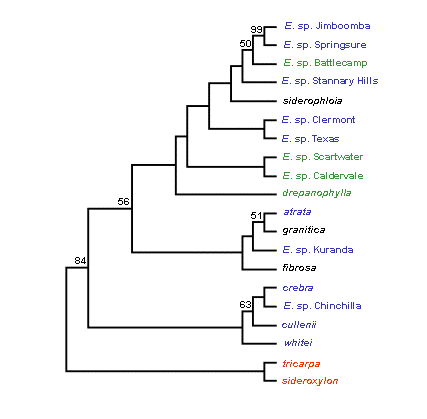
Figure 3. Fitch-Margoliash consensus trees without an evolutionary clock (left) and with an evolutionary clock (right) for eight formally recognised and 10 proposed new taxa in the Red Ironbark series Siderophloiae. Bootstrap percentages greater than 50% are shown at each branch. Colour used for taxon names refers to the Siderophloiae group shown in Table 1. Outgroup taxa are shown in red. [Note that the putative new taxa E. sp. Gilgandra and E. sp. Pentland are not presented. These formed close genetic alliances with E. sp. Chinchilla and E. sp. Stannary Hills respectively. This was concordant with morphological evidence indicating these two entities are not distinct from the latter two taxa and are now considered to represent range extensions (Tony Bean pers. comm.)].
A major finding of the study was support for the concept that E. crebra and E. drepanophylla have geographically restricted occurrences and that populations formerly assigned to these species represent new taxa identified by Tony Bean. The typology of proposed new taxa in both trees indicates they are genetically distinct. However, Siderophloiae taxa were characterised by high levels of allozyme variation and relatively similar allelic frequencies. This is reflected in the lack of bootstrap support for the clustering of most taxa in both trees. While the taxonomic recognition of proposed new taxa is supported, Bean’s informal grouping of taxa based on morphological attributes (shown in Table 1) was poorly resolved.
Strong genetic affinities are indicated for the grouping of ‘typical’ E. crebra with E. cullenii, E. whitei, E. sp. Chinchilla and the grouping of E. atrata, E. fibrosa, E. granitica and E. sp. Kuranda. These two alliances showed the same clustering patterns in both trees. Among other taxa, however, clustering patterns differed slightly in both trees. The Fitch-Margoliash consensus tree that assumed no evolutionary clock was the best fit for the informal grouping of taxa based on morphology, but there were some major genetic alliances among taxa that were not consistent with groupings based on morphology.
Taxa illustrating this trend include E. atrata and E. granitica. These clustered closely in both trees. However, based on morphological evidence E. granitica is not considered closely related to other taxa in the Siderophloiae (Hill and Johnson 1991). There was also evidence that E. sp. Kuranda is allied to E. granitica but based on morphological evidence it is placed in the ‘E. crebra group’ by Bean. Coincidentally, E. atrata, E. granitica and E. sp. Kuranda occur in relatively close geographic proximity and it is possible some gene flow may be occurring among these taxa. Another example of the lack of concordance between genetic and morphological groupings is provided by E. sp. Jimboomba and E. sp. Springsure from the ‘E. crebra group’. These taxa clustered with E. siderophloia from the ‘E. siderophloia group’ in both trees. The genetic affinities of taxa in the ‘E. drepanophylla group’ were also ambiguous and strong affinities to ‘E. crebra group’ taxa are indicated. More populations and taxa in the ‘E. drepanophylla group’ warrant survey before further conclusions regarding their phylogeny can be drawn.
Overall, the clustering patterns reflect a lack of concordance between genetic and morphological evidence used to group taxa at higher taxonomic levels. This result supports the systematic treatment of the Siderophloiae by Brooker (2000) who could not identify natural groups among taxa and placed all 22 recognised taxa in the subseries Subglaucae. The close genetic alliance among Siderophloiae taxa reflects the morphological trend of these taxa to have more similarities than differences. The recognition that taxa in the Red Ironbark series Siderophloiae represent a closely allied species-complex will have important implications for the domestication, harvesting and conservation management of these taxa. The new phylogeny of the Red ironbark series Siderophloiae will comprise at least 33 mainly tropical taxa, of which at least ten are yet to be formally described. Management of the genetic resources of this large important group will need to recognise that the group is characterised by relatively low levels of both genetic and morphological differentiation.
Acknowledgements
Many thanks to Maurice McDonald for his impeccable supervision
throughout the study.
Tony Bean, Charlie Bell, Penny Butcher and Ian Brooker are also thanked for
their input on various aspects of the project; the Centre for Plant Biodiversity
Research and CSIRO Forestry & Forest Products provided the scholarship
and resources.
References
Brooker MIH (2000). A new classification of the genus Eucalyptus L'Her. (Myrtaceae). Australian Systematic Botany 13: 79-148.
Felsenstein J (1995). PHYLIP (Phylogeny Inference Package) version 3.57. Distributed by the author. Department of Genetics, University of Washington, Seattle.
Hill, KD, Johnson LAS (1991). Systematic studies in the eucalypts - 4. New taxa in Eucalyptus (Myrtaceae). Telopea 4: 321-349.
Lewis PO, Zaykin D (2001). Genetic Data Analysis: Computer program for the analysis of allelic data. Version 1.0 (d16c).
Swofford DL, Selander RB (1989). BIOSYS-1: A computer program for the analysis of allelic variation in population genetics and biochemical systematics. Release 1.7. (University of Illinois, Department of Genetics: Illinois.)
Weir BS (1996). ‘Genetic Data Analysis II: Methods for Discrete Population Genetic Data.’ (Sinauer Associates: Sunderland, Massachusetts.)